As the first post on the STACCATO Early Stage Researcher’s (ESRs) Blog, I think it is a good idea to introduce the premise of what this Marie Skłodowska-Curie Actions (MSCA) funded project is all about and what my role is as one of its 11 ESRs/PhD Students.
Before continuing, I want to introduce what essentially underpins STACCATO, which is single cell analysis. The definition of a staccato is musical composition performed with each note sharply detached or separated from the others. To bring this into context with our project: We are separating and taking measurements from thousands of individual cells to study their intricate differences just like the notes in a staccato.
STACCATO is an initiative funded by the EU’s prestigious Marie Skłodowska-Curie Actions (MSCA) Innovative Training Network (ITN) programme. Prof Colin Clarke designed it to bolster Europe’s innovation capacity and leadership in the biopharmaceutical manufacturing sector.
Growing Europe’s pool of skilled scientists
If Europe’s biopharmaceutical sector is to continue to flourish and remain competitive in a booming international market, it is urgent that we invest in expanding our pool of highly skilled scientists and pioneer new analytical approaches to enable rapid design of efficient bioprocesses for biopharmaceutical production.
As an ITN, STACCATO has enabled the partnership of industry and academic experts from all over Europe to provide 11 ESRs (PhD Student researchers), of which I am one, with world-class intersectoral training and upskilling. This is to prepare us to tackle some of the most significant challenges facing the biopharma manufacturing industry.

One of the unique advantages of MSCA funded training networks is that ESRs have great opportunity to travel to partnered institutes for scientific collaboration throughout Europe, taking take part in training courses, presenting research at conferences and gaining invaluable industry experience during Secondments – extended international placements. I’m currently situated at Ireland’s National Institute for Bioprocessing Research and Training (NIBRT), but my first Secondment planned in 2020 will allow me to conduct some of my PhD research at the Paul-Ehrlich-Institut (PEI), another world-class institute for biomedicines research, in Langen, Germany.

What is single cell analysis and why do we care?
The underlying vision uniting the STACCATO ESRs, Principal Investigators and industry partners is to utilise high-resolution data captured from thousands of single cells to enhance and develop new manufacturing methods for biological medicines.
The majority of studies in cell biology are performed with large bulk numbers of cells, often over a million per sample. Although this is a powerful tool, by averaging over enormous cell populations we ignore the fact that no two cells are exactly alike even if they’re of the same type. Conventional bulk sample measurements overlook many important details that only become clear at single cell resolution. This could be likened to tasting a fruit smoothie and trying to identify everything in it; versus tasting each individual piece of fruit before everything was blended together.


Left Photo: by Tara Evans on Unsplash. Right Photo: by Rebecca Hansen on Unsplash
This ability to comprehensively profile the cellular heterogeneity of single cells is crucial to advancing our understanding of cell behaviours, disease mechanisms and identify previously elusive biomarkers and drug targets.
My project as a STACCATO ESR
Each ESR will focus on a different research aspect within STACCATO, such as developing single cell analysis methods or improving cell line performance, but all projects share the common goal to enhance bioprocessing development.
Here at NIBRT, I will be developing and applying methods for Single Cell Proteomics analysis to help in understanding the heterogeneity of industrially important cell lines utilized for biopharmaceutical manufacturing. The field of Proteomics encompasses all aspects of protein study, including protein identification, structural analysis, abundance and the qualitative and quantitative characterization of post translational modifications (PTMs). One of the most commonly used methods of doing this is called shotgun or bottom-up proteomics, whereby the proteins are first enzymatically digested into peptide fragments prior to separation and analysis by liquid chromatography-mass spectrometry (LC-MS), following this, specialised bioinformatics pipelines and vast databases are used to identify the proteins from their peptide sequences.
Mass Spectrometry warrants its own blog post, so I will go into detail on how this works and in relation to proteomics at a later time!
A science in its infancy
Single Cell Proteomics (SC-Proteomics) hasn’t quite been possible until recently, and it’s still in its infancy when compared to the powerful single cell RNA-sequencing methods that are used by thousands of labs around the world. SC-Proteomics is difficult because there is very little protein in a single cell (around 200 picograms!) and no equivalent of DNA amplification for proteins – a major bottleneck is losing what little protein you had throughout the sample preparation process.
Nikolai Slavov’s Lab at NEU Boston have developed SCoPE-MS (Single Cell ProtEomics by Mass Spectrometry), a method which tackles this issue by chemically labelling the individual cells and a ‘carrier channel’ of around 100 cells to absorb some of the protein losses during processing, which would otherwise be incurred by the single cell protein samples. However, there is still room for improvement here!
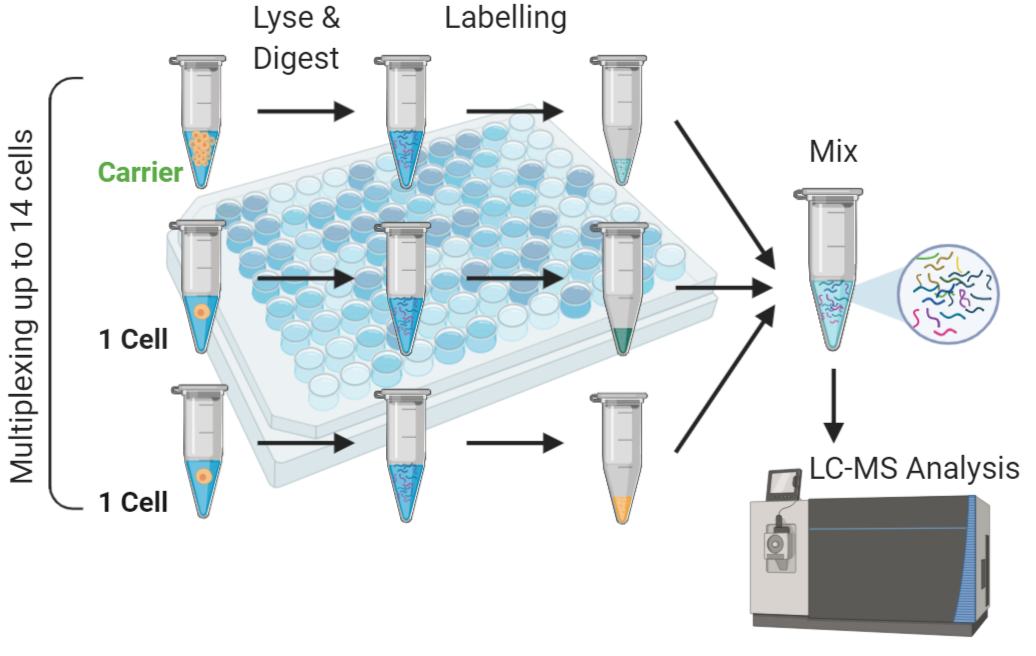
The different chemical labels of individual cells and carrier channel allows the relative quantification of protein for each cell to be seen in the MS data. I will apply similar methods to study CHO (Chinese Hamster ovary) cell heterogeneity, and attempt to make improvements in recovery of the cellular proteome and sample throughput (currently it takes around 10 days of instrument time to analyse around 1000 cells – that’s a long time!).
More to come
I hope that if you made it to the end of this; that you understand the gist of STACCATO, Single Cell Analysis and a little bit about my project at NIBRT. I will try to update here with posts that you may find interesting or educational on Mass Spectrometry, Single Cell Proteomics and gossip about the other ESRs. I am greatly looking forward to blog posts from the 10 talented and interesting people that I’ll be collaborating with over the next few years on the STACCATO Project.
Posted by Marco. R